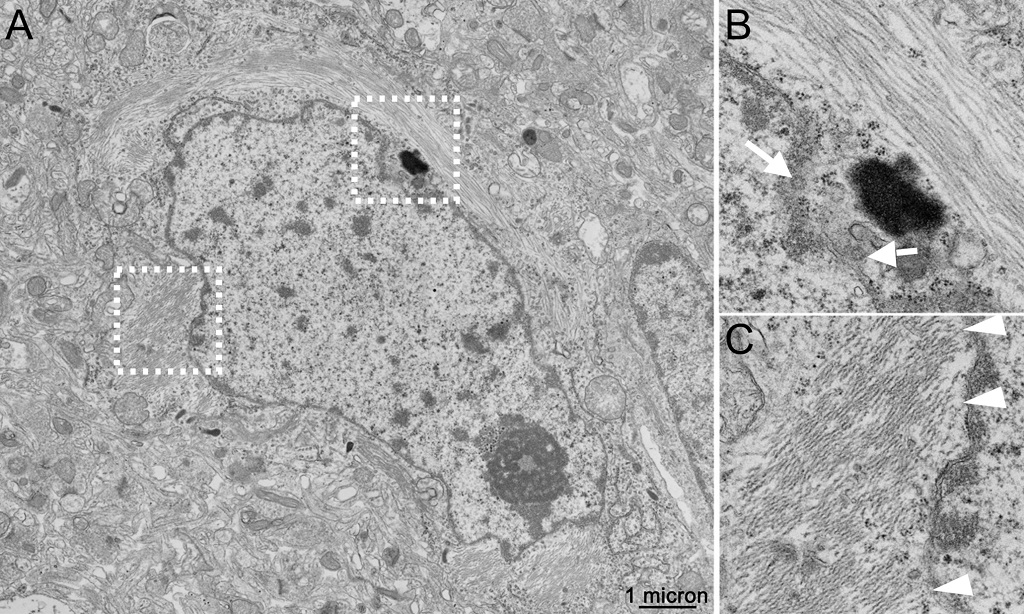
(A) Example transmission electron microscopy (TEM) micrograph of AD biopsy samples of cerebral cortex, showing excellent ultrastructural detail, including nuclear pores (white arrows in panel B, corresponding to white box in A) and microtubules, as well as examples of associations of paired helical filaments (PHF) with the membranes of the nuclear envelope (white arrowheads in panel C, corresponding to white box in A). The imaging studies which produced these data, originally conducted by us in the late 1980s, also highlighted associations of PHF with ribosomes, differences between the distribution of the Golgi apparatus in neurons containing PHF bundles, as compared to nearby and otherwise normal-appearing neurons without these filaments. Abnormalities in the microtubules were also documented in axonal processes that touched neuritic plaques.
A key outcome will be to deliver complete whole cell reconstructions of neurons and glia from unique biopsy samples of cerebral cortex taken from leading AD researchers in the 1960’s and 1980’s.1,2 We have focused on early onset cases, where cells effected to differing extents are neighbored by cells without AD-associated paired helical filaments (PHF), which is now considered to be largely made up of tau proteins. Many of these samples were screened and preliminarily reported on by Ellisman, Terry, and Mirra3 and later re-examined by us (Masliah and co-workers)4,5 and manifest near perfect preservation of ultrastructure, showing PHF and amyloid accumulations as well as modifications to subcellular organelles and cytoskeletons of the cell bodies, axonal and dendritic processes. Originally analyzed by us with early versions of serial section transmission EM reconstruction to produce small 3D data volumes,6 we propose to use very recent advances in automated 3D EM to provide a large reference collection of fully reconstructed brain cells as well as cell processes, ER and cytoskeletons in the vicinity of amyloid plaques.5 This deeper and more detailed analysis of these AD samples will exploit powerful deep learning-based approaches to rigorously and quantitatively extract detailed information about inter-organelle associations and cytoskeletal reorganization associated with neuronal degeneration of the type found in AD.