In recent work, we have extended our image segmentation capabilities to now include the use of a state-of-the-art deep learning convolutional neural network (CNN). This class-leading CNN, named DeepEM3D,85 was developed by Zeng and colleagues in the Dive Lab at Washington State University (Laboratory of Shuiwang Ji). Working closely with this group, we have constructed a pipeline to streamline the use of DeepEM3D at scale, pairing this CNN with pre- and post-processing operations to improve its performance for segmenting datasets (from both SBEM and EM tomography). Through participation in the NIH Data Commons Pilot program, we have also developed and have released (in an open distribution) an Amazon Machine Image (AMI) of this now integrated and richly engineered software suite, which simplifies the installation and execution of this software on commercial cloud GPU nodes (per NIH’s cloud credit model).86 We have also developed a docker container to similarly streamline deployment and execution on National XSEDE cluster resources (e.g., the Comet cluster for which we will enjoy a large dedicated allocation of compute cycles to perform this work – See letter of support from Michael Norman, Director of the San Diego Supercomputer Center). We call this fully productionized and cloud/cluster deployable implementation –
CDeep3M.
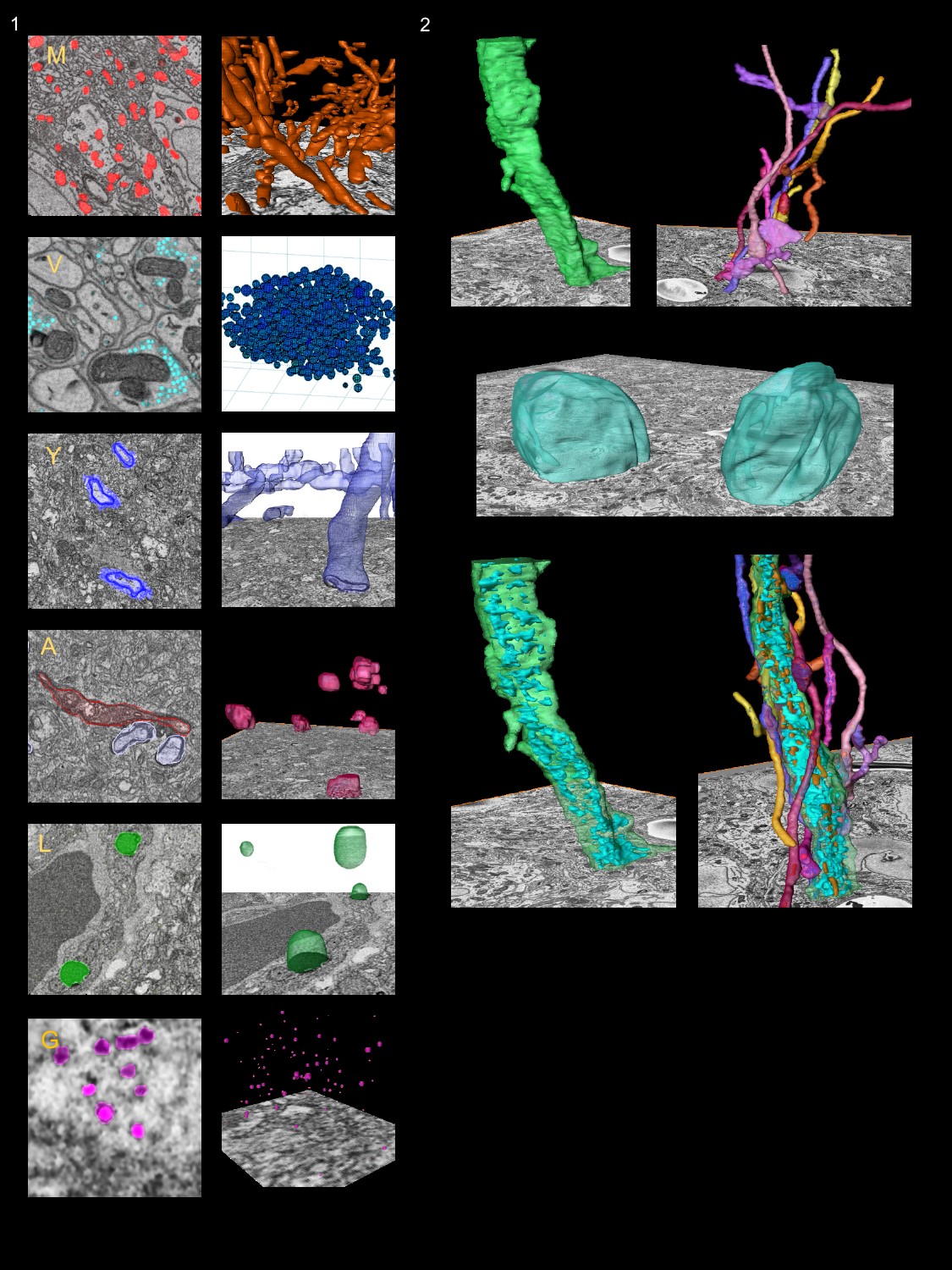
Leveraging this body of work, we will perform image segmentation of representative AD reference volumes, targeting key structures, including mitochondria, synaptic vesicles, Golgi apparatus, endoplasmic reticulum, lipofuscin, and myelin. These segmentations will be bundled with the raw data, ground truth training data, and CNNs (trained to these target structures) for ready access by the community.
To maximize the impact of the data collected and the effort (and computational cycles) expended to produce quantitative data products, we will immediately share all data volumes and intermediates, derived CNN segmentation models, related training data, and the tools used in the processing of these data with the AD community. Our goal is to not only share this valuable data but also facilitate the continued harvesting of new information from these massive scenes with scalable, extensible tools that can be invoked by any research scientist with a web browser and an Amazon Web Services cloud computing account.
Learn more about CDeep3M